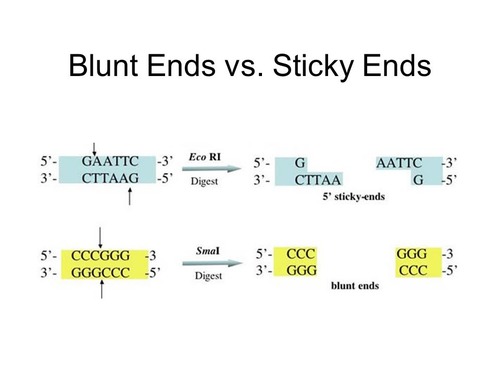
Restriction enzymes derived from bacteria are used in recombinant DNA techniques. The enzyme BamHI cuts DNA in the sequence
G↓
G─A─T─C─C
C─C─T─A─G ↑
G
When used to cut the sequence
5’…ATGGATCCGGACTAA…3’
3’…TACCTAGGCCGGATT…5’
How many DNA fragments are formed?
A) 2 with flat ends
B) 3 with cohesive ends
C) No fragments are formed
D) 2 with cohesive ends
E) 3 with flat ends
The answer is D. Could anyone help?